SNP PolTaq DNA Polymerase 2X PCR master mix
Order number: M3128.0100
Shipping: Shipment: on wet ice. Store at -20°C. For laboratory usage only! Ready to ship today,
Delivery time 1-3 workdays
Short shelf life. Available only while supplies last!
*Prices plus VAT plus shipping costs
The SNP PolTaq DNA polymerase used for the SNP PolTaq 2X Master Mix has been specially designed for easy, reliable and rapid allele-specific discrimination, eg. CRISPR / Cas9 point mutations, for detecting incorrect CRISPR / Cas9 products, or validating sequencing results. The SNP PolTaq DNA polymerase distinguishes highly specific, whether a mismatch of the primer-template-complex is present or not. The mismatch (point mutation) must be at the 3 'end of the primer. Thus, mutant alleles can be distinguished exactly from wild-type alleles - without sequencing, since the polymerase simply does not amplify in the case of a mismatch.
Just place your primer on the supposed point mutation (Important: The point mutation must be at the 3 'end) and the polymerase will detect a mismatch in this region with almost 100% accuracy: If the template base complementary to the 3' end of the primer shows the mutation and the primer does not, no amplification takes place - 100% certainty within a short time!
The SNP PolTaq DNA polymerase can therefore be easily, time and cost-effectively used for the screening of point mutations.
The SNP PolTaq DNA polymerase has 5'-3 'nuclease activity and can therefore be used for specific hydrolysi probes such as Taqman® probes or Molecular beacons.
For more information on our SNP DNA polymerases and applications, read our blog now.
Test sample available at a special price! No shipment costs within Germany. The test sample price will be refunded on the first official order of the product.
Description
SNP PolTaq DNA polymerase is a highly selective DNA polymerase for the detection of Single Nucleotide Polymorphisms. It was developed specifically for allele-specific discrimination, where a very high discrimination rate is required e.g., in allele-specific PCR (ASA; AS-PCR), allele-specific primer extension (AS-PEX), SNP analysis, genotyping or in methylation-specific PCRs (MSP). Many other DNA polymerases tolerate mismatched primer-template complexes and are therefor not suitable. The SNP PolTaq DNA polymerase, on the other hand, distinguishes these specifically (high discrimination) and supplies only PCR products with perfectly matching primer pairs! The Genaxxon SNP Pol and SNP PolTaq DNA polymerases differ up to 100% by means of allele-specific PCR between the two alleles and, after a simple qPCR, give a clear result as to which allele is present. Thus, the principly great potential of the CRISPR/Cas9 technology can be used for human medicine and plant biotechnology especially together with the SNP PolTaq or SNP Pol DNA polymerase.
Allele-specific PCR can be used to quantify the mutation rate in a pool or background of wild-type sequences. The verification of mutation frequencies determined by NGS can also be verified by means of allele-specific PCR and SNP Pol DNA polymerase. The SNP PolTaq DNA polymerase is very suitable for the analysis of liquid biopsy samples. With SNP PolTaq, the presence and frequency of cancer mutations can be analyzed and quantified very well.
Picture below: Application note SNP Pol DNA Polymerase
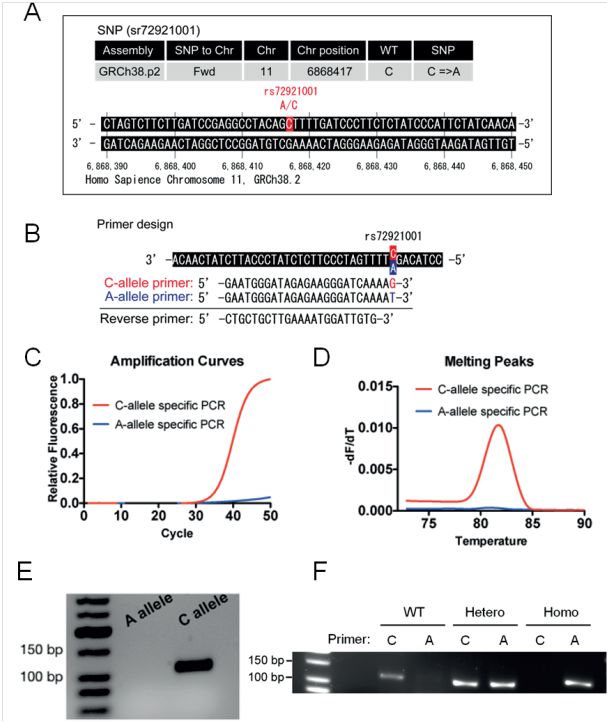
We recommend short amplicon length (about 60-200 bp) for best results, but also longer amplicon lengths are possible. The addition of additional Magnesium (+0.5 - 1.5mM) might be needed in case of longer amplicons >500 bp.
The SNP Pol DNA polymerase (M3009 > or M3061 >) can be used together with non-specific fluorescent dyes (eg Genaxxon's Green DNA Dye > or SybrGreen®) in real-time PCR. When working with specific PCR probes, only the SNP PolTaq DNA polymerase can be used, since only these have a 5'-3 'exonuclease activity.
With our high quality dNTPs as Set (M3015.4100) > or Mix (M3016.1010) > or our DNA Ladders > and our favourable standard agarose (M3044) > we can offer additional products for your PCR.
Application areas for SNP Pol DNA and SNP PolTaq DNA polymerase
- Monitoring, verification and detection of point mutations
- Identification of correct or wrong CRISPR/Cas9 products
- Verification/validation of sequencing results
- Quantification of mutations (e.g. NGS results)
- SNP-detection by allele-specific amplification (ASA) / Allele-specific PCR
- Methylation specific PCRs (MSP) after bisulfite treated DNA (CpG methylation sides)
- HLA genotyping
- micro sequencing
- realtime PCR with hydrolysis probes
- realtime multiplex PCRs
- DamID-seq data in C. elegans.
As an alternative to ChIP, DNA adenine methyltransferase identification by sequencing (DamID-seq) was recently shown to be able to characterize binding sites in single mammalian cells. Additionally, DamID can be achieved for cell-type specific analysis by expressing Dam fusion proteins under tissue specific promoters in a controlled manner. In this report, we present a user-friendly pipeline to analyse DamID-seq data in C. elegans.
Sharma R, Ritler D, Meister P.
Tools for DNA adenine methyltransferase identification analysis of nuclear organization during C. elegans development. Genesis. 2016 Feb 4. doi: 10.1002/dvg.22925.
- Minisequencing SNP genotyping with SNPase DNA Polymerase can be carried out by the procedure described in:
Lovmar L, Fredriksson M, Liljedahl U, Sigurdsson S, Syvänen AC.
Quantitative evaluation by minisequencing and microarrays reveals accurate multiplexed SNP genotyping of whole genome amplified DNA. Nucleic Acids Res. 2003;31:e129.
- Allel specific mismatch selectivity by the HiDi DNA polymerase
high single nucleotide discrimination SNP
Technical Data:
Specifications:
2-time master mix. Contains all components necessary for direct PCR or PCR based genotyping.
SNP PolTaq DNA polymerase shows 5’-3’ exonuclease activity! Especially applicable for usage together with hydrolysis probes like, e.g. TagMan™ probes.
application:
- CRISPR/Cas9 controls - validation of NGS sequencing results - SNP-detection by allele-specific amplification (ASA) / Allele-specific PCR - allelspezfische primer extension (AS-PEX) - Methylation specific PCR (MSP) - HLA genotyping - Micro-sequencingSource
rec. from E.coliSicherheits Hinweise / Safety
Klassifizierungen / Classification
eclass-Nr: 32-16-05-02Dokumente - Protokolle - Downloads
Here you will find information and further literature on SNP PolTaq DNA Polymerase 2X PCR master mix. For further documents (certificates with additional lot numbers, safety data sheets in other languages, further product information) please contact Genaxxon biosience at: info@genaxxon.com or phone: +49 731 3608 123.
Dokumente - Protokolle - Downloads
Kann man die SNP TaqPol DNA-Polymerase mit Bisulfit-behandelten DNA-Proben verwenden?
Ja, unsere SNP TaqPol DNA-Polymerase ist beispielsweise hervorragend geeignet für die methylierungsspezifische PCR (MSP). Da eine einzelne Fehlpaarung für bestimmte MSP-Ergebnisse ausreicht, ermöglicht SNP TaqPol DNA-Polymerase sogar eine zuverlässige MSP-Analyse einzelner CpG-Stellen.
Kann man SNP TaqPol 2x PCR Master Mix und SNP TaqPol DNA-Polymerase in auf Sonden basierenden Assays verwenden?
Ja, das ist möglich. Die SNP TaqPol DNA-Polymerase besitzt 5'-3'-Nuklease-Aktivität und kann daher auch in hydrolysebasierten Sondenassays (z. B. TaqMan®) verwendet werden.
Was ist die Fehlerrate von SNP PolTaq DNA-Polymerase?
Die Fehlerrate von SNP TaqPol ist vergleichbar mit der Fehlerrate von Wildtyp-Taq DNA-Polymerase.
Kann man SNP PolTaq DNA-Polymerase in Real Time-PCRs mit einem qPCR-Farbstoff wie SYBR®Green verwenden?
Ja, unsere SNP PolTaq DNA-Polymerase kann auch zusammen mit SYBR®Green verwendet werden. Es ist aber darauf zu achten, dass die SYBR®Green-Konzentration nicht das 0,1-fache übersteigt, da es sonst zur Inhibition der PCR kommt (Normalerweise wird SYBRGreen als 1-fach Konzentrierte Menge in der PCR eingesetzt!).
Für welche Amplikonlänge kann die SNP PolTaq und die SNP Pol DNA-Polymerase benutzt werden?
Wir empfehlen eine Amplikonlänge von ca. 60-200 bp, um optimale Ergebnisse zu erzielen. Es sind jedoch auch größere Amplikonlängen möglich.