Since the introduction of FAST qPCR, the world of real-time PCR/qPCR has entered a revolutionary era. By using special instruments and optimized master mixes aimed at speed and efficiency, this technology allows you to accelerate your qPCR applications without compromising on specificity and sensitivity.
Real-time PCR, also known as qPCR, is undoubtedly an impressive method for measuring the amplification of target DNA in real time. However, some people may despair at the abundance of terms, protocols and kits on the market. This begs the question: are reliable qPCR results only achievable through magic or is it ultimately more about the art of precise fine-tuning?
We can reassure you: in fact, with just a few tricks you can optimize your qPCR so that you can easily achieve reliable and accurate results. Whether you are already an expert in the field of qPCR or just taking your first steps into this fascinating world, this article will guide you through the key factors that will ensure your qPCR assays produce the best possible results. From deciding on the method, to quality control, to optimizing specificity - we have important tips and suggestions for you.
We can reassure you: in fact, with just a few tricks you can optimize your qPCR so that you can easily achieve reliable and accurate results. Whether you are already an expert in the field of qPCR or just taking your first steps into this fascinating world, this article will guide you through the key factors that will ensure your qPCR assays produce the best possible results. From deciding on the method, to quality control, to optimizing specificity - we have important tips and suggestions for you.
Problems with non-specific bands after your PCR? Or, the PCR does not work, but you find lots of amplified primer dimers? No desire for hectic and cooling when pipetting your PCR and the following steps? Looking for reproducible results summer and winter? Then simply use a hot-start PCR.
Maybe you already know it: Even if you did not make a mistake when pipetting your PCR, the results can sometimes be frustrating and disappointing. But why? Conventional DNA polymerases are already active at room temperature - and this can lead to many "falsely amplified" templates, false-positive results and low yield.
Maybe you already know it: Even if you did not make a mistake when pipetting your PCR, the results can sometimes be frustrating and disappointing. But why? Conventional DNA polymerases are already active at room temperature - and this can lead to many "falsely amplified" templates, false-positive results and low yield.
The world of genomics and molecular biology has taken a breathtaking leap into the future in recent years. In addition to "high fidelity" enzymes, "highly discriminating" enzymes such as the highly selective SNP DNA polymerases from Genaxxon have become the superstars of the laboratory. These enzymes are specifically modified for highly selective PCR tests and mutation detection - they help you when you want to find "the needle in the haystack".
Since the isolation of the first Taq DNA polymerase in 1976, PCR has become an integral part of the molecular biology laboratory. Of course, methods, reagents and protocols have evolved. The goal is clear: to make it better, faster, cheaper, and easier. Every day, lab technicians, students and scientists around the world spend countless hours pipetting and preparing PCR experiments. This makes it even more frustrating when errors and contamination only become apparent in the final step of the PCR workflow.
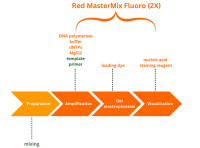
But how can the work in the PCR laboratory be optimised to ultimately minimise sources of error and at the same time save time, money, and nerves? We at Genaxxon would like to shed some light into the darkness of the laboratory and show how our new products around the Red Mastermix Fluoro (2X) can help to optimise your PCR workflow along a typical PCR workflow.
But how can the work in the PCR laboratory be optimised to ultimately minimise sources of error and at the same time save time, money, and nerves? We at Genaxxon would like to shed some light into the darkness of the laboratory and show how our new products around the Red Mastermix Fluoro (2X) can help to optimise your PCR workflow along a typical PCR workflow.
Do you already work with multiplex qPCR? And do you appreciate the error, cost and time savings of the multiplex qPCR method? But does your multiplex assay already deliver the best possible results? Specific quality and service features are crucial for you to perform multiplex qPCR accurately and, above all, conveniently. Thanks to the newly developed Taq DNA polymerase in our 5X qPCR Multiplex MasterMix, Genaxxon bioscience offers a solution that guarantees robust PCR performance for a wide range of different applications.
Have you been interrupted while pipetting and forget which well you have already pipetted into? Pipetting loading buffer takes too long? The DNA sample is not completely clean? Then use Genaxxon's Red MasterMix for your PCR - guaranteed high specificity for the best results in your lab. Here are 4 good reasons why you should use Genaxxon's Red MasterMix to simplify your lab work without sacrificing precision.
How to save money using SafeGel from Genaxxon as DNA staining dye for gel electrophoresis
Everything seemed to be easier in the past. At least that’s true when it comes to staining DNA in agarose gels. Ethidium bromide was the agent of choice, despite its mutagenic effect and costly disposal. Today, one is spoilt for choice. But which DNA staining dye is the right one? GelRed® as an alternative is non-toxic and has a high sensitivity, but the price makes one cringe.
Everything seemed to be easier in the past. At least that’s true when it comes to staining DNA in agarose gels. Ethidium bromide was the agent of choice, despite its mutagenic effect and costly disposal. Today, one is spoilt for choice. But which DNA staining dye is the right one? GelRed® as an alternative is non-toxic and has a high sensitivity, but the price makes one cringe.
Progress in the field of SNP genotyping and its applications, from the perspective of plant breeding and agriculture
Sanger sequencing technology was the first generation of sequencing and helped provide basic knowledge about genome sequencing but didn’t have the potential for its further improvement. Hence a new technological jump in sequencing expertise was required. That’s when Next Generation Sequencing techniques came into existence and revolutionized our entire capability of genome sequencing.
Extraction and isolation of viral RNA is the first step in the detection and testing of viral RNA by real-time RT-PCR. Here we compare different methods and protocols for extraction of viral RNA.
Nested RT-PCR and real-time RT-PCR are the most common methods for the analysis of viral infections (see Robert Koch Institute >). In contrast to examinations with ELISA or IFA, this can be done at an earlier stage of infection.
Nested RT-PCR and real-time RT-PCR are the most common methods for the analysis of viral infections (see Robert Koch Institute >). In contrast to examinations with ELISA or IFA, this can be done at an earlier stage of infection.
Non-protein-coding RNAs (ncRNAs) are expressed in viruses, archaea, prokaryotes and eukaryotes. They fulfil vital roles in the regulation of chromatin architecture, epigenetic memory, transcription, RNA splicing, editing and turnover. To date, at least 18 distinct types of ncRNA have been identified, which are classified according to their size, function, subcellular localization and target specificity.
In the articles below you will find the authors already relying on the Genaxxon products. We say thank you to these authors, trusting in our service. Let us convince you as well.
Wine production, while considered an art form, owes an increasing debt to the latest molecular genetic technologies. Not only do these facilitate batch-to-batch consistency, they also enable the generation of novel strains of grape microflora. While the choice of grapes is important, so too is the interplay between extrinsic and intrinsic factors, and the technological decisions made at every step in the process. Together, these determine whether the finished product will be classed as a “vintage wine”, a “quaffing wine”, or “vinegar”.
What are the differences between proteins, peptides and amino acids and and how did these molecules originally come about?
SNP PolTaq DNA polymerase for better Sake taste and flavor: SNP PolTaq for qPCR for the unambiguous identification of point mutations in yeast!
Read more about a recent case report from Japan.
Read more about a recent case report from Japan.
Nowadays there are many different suppliers and systems for polymerase chain reaction (PCR) and of its derivative techniques, quantitative, or real-time, PCR (qPCR) and reverse transcription PCR (RT-PCR). Competition gives you the opportunity to simplify diagnostics and to make research more financially favorable.
We are proud to inform you that Genaxxon bioscience has been successfully certified according TÜV ISO 9001: 2015!
There's no accounting for taste.
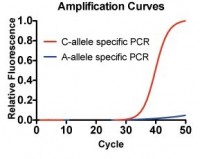
Cilantro (Coriander): some people love it in their food, some hate it. Here we are detecting a genomic SNP (rs72921001) in HeLa genomic DNA. This SNP is reported to be close to a number of genes coding for olfactory receptors. (Reference: Eriksson N. et al. (2012), "A genetic variant near olfactory receptor genes influences cilantro preference.")
Considering, that only the C-allele specific primer is extended and yielding in a specific amplicon, we can conclude a genetic predisposition in disliking cilantro, as this SNP is significantly associated with detecting a soapy taste to cilantro.
Cilantro (Coriander): some people love it in their food, some hate it. Here we are detecting a genomic SNP (rs72921001) in HeLa genomic DNA. This SNP is reported to be close to a number of genes coding for olfactory receptors. (Reference: Eriksson N. et al. (2012), "A genetic variant near olfactory receptor genes influences cilantro preference.")
Considering, that only the C-allele specific primer is extended and yielding in a specific amplicon, we can conclude a genetic predisposition in disliking cilantro, as this SNP is significantly associated with detecting a soapy taste to cilantro.