SNP PolTaq DNA polymerase can help you to identify mutations in yeast - for better sake taste and flavour.
SNP Pol DNA polymerase > and SNP PolTaq DNA polymerase >, developed for their ability to identify point mutations with greater specificity and efficiency than other commercially available enzymes, are currently the gold standard for use in allele-specific mutation analysis, in a range of applications.
A case report from Japan: Whether for the making of beer, wine or sake, different yeasts create different tastes in the final product, due to factors such as how the sugar-to-alcohol conversion is carried out and the types of by-products that are released. Two types of sake, considered especially high-quality, are daiginjo-shu and junmai-daiginjo-shu. Both are often made using the yeast strain K1801. A previously-identified mutation in K1801 was recognised as a desirable change that makes the yeast produce high amounts of ethyl caproate, the chemical that creates a fruity taste in many varieties of high-quality sake. Read more from Asian Scientist Magazine at: https://www.asianscientist.com/2016/07/in-the-lab/gene-mutation-japanese-rice-wine-sake/
In the brewing of both sake and beer, the saccharification of starch is catalysed by enzymes in the mould, Kôji, while yeast enzymes mediate alcoholic fermentation. These two processes occur sequentially in beer fermentation, but remarkably, in sake brewing, the two processes take place simultaneously and continuously. As the Japanese market does not accept sake made from genetically-modified yeast, the next step for the research team is to screen potentially thousands of K1801 yeast cells until a naturally-occurring clone with the desired mutation(s) can be identified and isolated.
This is where SNP PolTaq DNA polymerase > comes into play and can make a big difference. Many publications have shown that allele-specific PCR amplification using SNP PolTaq Polymerase can reliably improve the positive identification of yeast mutants that produce sake with better taste and flavor. SNP PolTaq DNA polymerase shows 5'-3'-nuclease activity, and is therefore suitable for hydrolysis probe-based qPCR assays (Taqman®, molecular beacons, etc.).
SNP Pol DNA polymerases > are highly selective DNA polymerase variants. They have been selected for use in assays in which High Discrimination is required, for example, in allele-specific, or methylation-specific PCRs. Whereas many DNA polymerases tolerate mismatched primer-template complexes, SNP Pol DNA polymerase efficiently identifies these and produces specific amplicons only when the primer pairs are matched perfectly. This makes them indispensable for mutation detections, SNP detections, HLA genotyping or the analysis of single CpG methylation sites.
Application areas for SNP PolTaq DNA polymerase
- Control and detection of point mutations
- Identification of the „right“ clones
- Control and validation of sequencing results
SNP Pol DNA polymerase can be applied to allele-specific PCR of DNA templates from diverse organisms
Below are data kindly provided from a customer at the University of Konstanz (research group of Prof. Dr Kroth), who successfully used SNP Pol DNA polymerase to identify indels by allele specific PCR of microalgae (Phaeodactylum tricornutum):

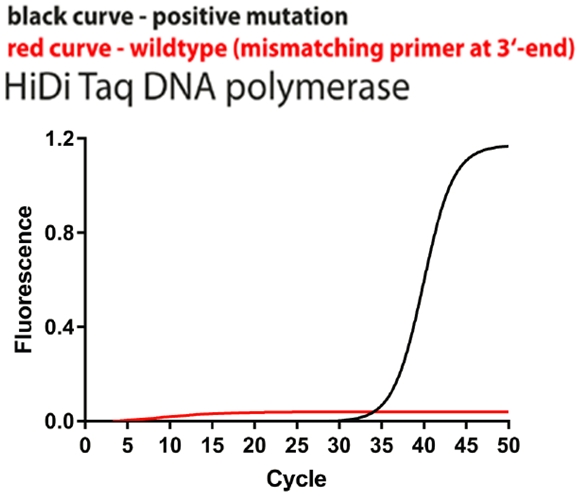
In our previous blog > we provided evidence of the greater target specificity of SNP PolTaq DNA polymerase, relative to that of other DNA polymerases. Here, we show that SNP Pol DNA polymerase is considerably less likely than other, comparable DNA polymerases, to amplify templates that exhibit a 3’-terminal mismatch with the primer.
Features of SNP Pol
Fast (shortened): Requires only the time needed for a real-time PCR. This is significantly faster than sending a plasmid for sequencing, waiting for the sequencing result and subsequent evaluation of the sequencing results, etc.
Easy (without laborious sequencing): PCR is an established procedure in almost every laboratory. Fast: carried out in your own (or in a neighboring) laboratory. Feasible: Normally, PCR is available anyway. No shipping, no evaluation of sequencing results.
Cost-effective: Cheaper than sequencing each individual sample. A Sanger sequencing sample costs approximately between 2.50-7.50 €, a PCR reaction only a few euro cents.
“To identify yeast mutants with a point mutation, detection of the specific mutant alleles is necessary. For this purpose, we applied allele-specific polymerase chain reaction (PCR) to detect the FAS2-1250S dominant mutant allele that encodes an altered fatty acid synthase in Japanese brewer's yeast strains. These strains are known to produce a higher amount of ethyl caproate in Japanese sake. The mutant strains were supposed to be diploid and to contain heterozygous alleles, including wild-type FAS2 and a dominant FAS2-1250S. A set of oligonucleotide primers was designed to contain different nucleotides at their 3′ termini: one type was identical to the wild type and the other to the mutant FAS2. Another set of primers was designed to have an additional mismatch at the second nucleotide from their 3′ termini. By testing with control strains, we established PCR conditions for specific amplification. Using these conditions and a simple template preparation procedure with SDS, the presence of the allele was detected in commercially used sake yeast strains. The method presented here will be useful for the identification of specific yeast strains.”
References:
- Goshima T, et al. (2016) Identification of a mutation causing a defective spindle assembly checkpoint in high ethyl caproate-producing sake yeast strain K1801. Biosci Biotechnol Biochem 80(8):1657-62
- Akada R, et al. (2001) Detection of a point mutation in FAS2 gene of sake yeast strains by allele-specific PCR amplification
- DamID-seq data in C. elegans / Sharma R, Ritler D, Meister P.
- Minisequencing SNP genotyping with SNPase DNA Polymerase / Lovmar L, Fredriksson M, Liljedahl U, Sigurdsson S, Syvänen AC
- Allele specific mismatch selectivity by the HiDi DNA polymerase / Drum M, Kranaster R, Ewald C, Blasczyk R, Marx A