SNP analysis
SNPs (spell snips) have been identified in all genomes and can be used for a multitude of analyses, including studying mutations implicated in various cancers, genetic disease research, mitochondrial DNA investigations, scrapie susceptibility in sheep, loss of heterozygosity, assessing performance in food animal production, and even differentiating drug and non-drug forms of Cannabis.
The Genaxxon SNP DNA polymerase > is a highly selective DNA polymerase variant, specially evolved for allel specific discrimination, for instance in allele-specific PCRs (ASA), primer extensions or methylation-specific PCRs (MSP), where our SNP DNA Polymerase shows excellent results.
The Genaxxon SNP DNA polymerase > efficiently discriminates primers which have only one mismatch at the 3'-end whereas many other DNA polymerases tolerate mismatched primer-template complexes. SNP DNA polymerase > efficiently discriminates those and only produces specific amplicons in case of perfectly matched primer pairs. This makes SNP Pol DNA polymerase very useful for SNP detections, HLA genotyping, the analysis of single CpG methylation sites or multiplexing PCR assays!
For realtime PCR with or without Probes we offer a “Taq SNP polymerase variant > of our SNP Pol DNA Polymerase, which shows 5'-3'-nuclease activity and therefore can be used for hydrolysis probe-based assays together with Molecular beacons or TaqMan® probes
SNP genotyping identifies single nucleotide polymorphisms (SNPs) that are common DNA variants present across the human genome. SNPs have been shown to be responsible for differences in genetic traits, susceptibility to disease, and response to drug therapies.
Genotyping of SNPs has become extremely important to researchers working to understand and treat disease. SNPs occur approximately once every 100 to 300 bases and can be detected by various different techniques such as RFLP, SSCP, sequencing, allele specific PCR and more.
A wide range of human diseases, e.g. sickle-cell anemia, β-thalassemia and cystic fibrosis result from SNPs. The severity of illness and the way our body responds to treatments are also manifestations of genetic variations. A single SNP may cause a Mendelian disease, e.g. a single base mutation in the APOE (apolipoprotein E) gene is associated with a higher risk for Alzheimer's disease though for complex diseases, SNPs do not usually function individually, rather, they work in coordination with other SNPs to manifest a disease condition as has been seen in Osteoporosis.
SNPs' greatest importance in biomedical research is for comparing regions of the genome between cohorts (such as with matched cohorts with and without a disease) in genome-wide association studies >. SNPs have been used in genome-wide association studies as high-resolution markers in gene mapping related to diseases or normal traits. SNPs without an observable impact on the phenotype (so called silent mutations) are still useful as genetic markers in genome-wide association studies, because of their quantity and the stable inheritance over generations.
The knowledge of SNPs will help in understanding pharmacokinetics (PK) or pharmacodynamics, i.e. how drugs act in individuals with different genetic variants. Diseases with different SNPs may become relevant pharmacogenomic targets for drug therapy. Some SNPs are associated with the metabolism of different drugs.
Example
Cilantro (Coriander):
There's no accounting for taste!
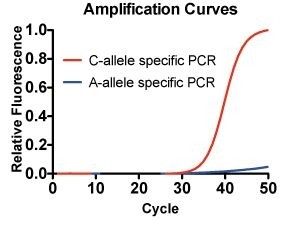
Cilantro: some people love it in their food, some hate it. Here we are detecting a genomic SNP (rs72921001) in HeLa genomic DNA. This SNP is reported to be close to a number of genes coding for olfactory receptors. (Reference: Eriksson N. et al. (2012), "A genetic variant near olfactory receptor genes influences cilantro preference.").
Considering, that only the C-allele specific primer is extended and yielding in a specific amplicon, we can conclude a genetic predisposition in disliking cilantro, as this SNP is significantly associated with detecting a soapy taste to cilantro.
Allele-specific PCRs were performed from 1ng/µL of HeLa gDNA in the presence of a realtime dye, indicating the amplification of the C-allele specific primer only. The A-allele specific primer is discriminated, thus not amplified up to 50 cycles.
PCR products were subsequently analysed on a 2.5% agarose gel. A specific product is visualized by ethidium bromide staining at the right amplicon length of 109 bp.
SNP PolTaq DNA polymerase > efficiently amplifies from primers that are matched at the 3'-end and discriminates primers that are mismatched.